Example Projects
Antibody structure prediction using deep learning
Brennan Abanades Kenyon
Supervisor: Professor Charlotte M. Deane
Industrial Supervisors: Guy Georges (Roche) and Alexander Bujotzek (UCB)
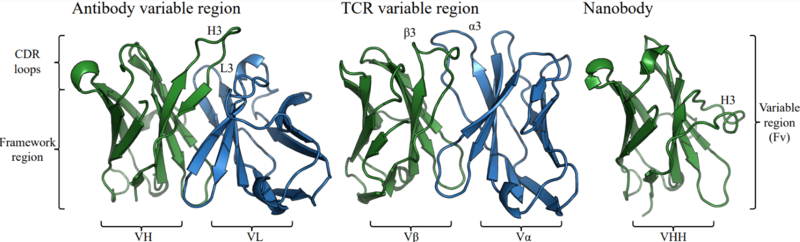
Structural representation of an antibody variable domain (PDB code 1GIG), a TCR variable domain (PDB code 7SU9) and a nanobody (PDB code 4LAJ) with labelled regions.
The aim of my doctoral project is to improve the prediction of antibody structures and optimise the use of existing antibody data. As a starting point, we developed ABlooper [1], a tool that predicts the position of backbone atoms for CDR loops in antibodies. This was the first method to incorporate equivariant deep learning into antibody structure prediction. However, ABlooper has some limitations: it cannot model side-chain atoms and sometimes generates unphysical structures, which need to be corrected using computationally expensive methods.
Our second iteration for antibody structure prediction, ABodyBuilder2 [2], addresses some of these challenges. Inspired by the AlphaFold2 [3] model, ABodyBuilder2 directly predicts side chain atoms, thus enhancing the accuracy of their modelling. By incorporating hard-coded rules for bond angles and distances within a residue, it also reduces the number of unphysical structures predicted. ABodyBuilder2 is as accurate as AlphaFold-Multimer [4] while achieving over a hundred-fold increase in speed. At the time of release, it stands as the most accurate of all antibody-specific methods. When we trained the model exclusively on T-Cell Receptors (TCRs) and nanobodies, comparable results were obtained, leading to the creation of TCRBuilder2 and NanoBodyBuilder2.
Concurrently, we introduced two innovative tools: KA-Search [5] and PLAbDab [6], designed to effectively harness publicly available antibody data. KA-Search facilitates rapid and exhaustive searches in vast NGS antibody databases based on sequence identity. PLAbDab compiles patent and paper-derived antibody sequences into a searchable database and makes them searchable either by sequence or structure. We anticipate that these tools, alongside our improved structural modelling, will aid in the process therapeutic antibody design.
All methods developed during this PhD are freely available either as web servers (https://opig.stats.ox.ac.uk/webapps/sabdab-sabpred/sabpred) or on GitHub (https://github.com/oxpig/).
[1] Abanades, Brennan, et al. “ABlooper: Fast accurate antibody CDR loop structure prediction with accuracy estimation”. In: Bioinformatics 38.7 (2022), pp. 1877–188
[2] Abanades, Brennan, et al. “ImmuneBuilder: Deep-Learning models for predicting the structures of immune proteins.” In: Communications Biology 6.1 (2023), p. 575
[3] Jumper, John, et al. "Highly accurate protein structure prediction with AlphaFold." Nature 596.7873 (2021), pp. 583-589.
[4] Evans, Richard, et al. "Protein complex prediction with AlphaFold-Multimer." bioRxiv (2021)
[5] Olsen, Tobias H., et al. “KA-Search: Rapid and exhaustive sequence identity search of known antibodies”. In: Scientific Reports 13.1 (2023), p. 11612.
[6] Abanades, Brennan, et al. “The Patent and Literature Antibody Database (PLAbDab): an evolving reference set of functionally diverse, literature-annotated antibody sequences and structures” In: bioRxiv (2023).
PKPD modelling: Towards personalising treatments in clinical practice
David Augustin
Supervisors: David Gavaghan, Ben Lambert, Martin Robinson
Industrial supervisors: Ken Wang (Roche), Antje-Christine Walz (Roche)

The implementation of framework for the use case of warfarin treatment
David’s research focus is the study of treatment response variability and modelling approaches to mitigate it. Treatment response variability across patients is a common phenomenon in clinical practice. For many drugs this inter-individual variability does not require much (if any) individualisation of dosing strategies. However, for some drugs, including chemotherapies and some monoclonal antibody treatments, individualisation of dosages are needed to avoid harmful adverse events. Model-informed precision dosing (MIPD) is an emerging approach to guide the individualisation of dosing regimens of otherwise difficult-to-administer drugs.
Several MIPD approaches have been suggested to predict dosing strategies, including regression, reinforcement learning (RL) and pharmacokinetic and pharmacodynamic (PKPD) modelling, but a unified framework to study the strengths and limitations of these approaches was missing. David developed a framework for the simulation of clinical MIPD trials [1].
In the context of this project, David developed the open-source Python package Chi which can be used to implement and simulate treatment response models [2]. The package simplifies the reproduction of clinical trial simulation and enables researchers to implement their own clinical trial models. In addition, Chi integrates with the open-source Python package Pints which makes the estimation and inference of model parameters from data possible [3].
A central research theme in David’s work is the quantification of model uncertainty, including parametric and structural uncertainty. In a recent publication, David introduces a novel method for the estimation of the parameters of nonlinear mixed effects models from snapshot measurements [4]. In another publication, David discusses the limitations of information criteria-based model selection for the estimation of structural uncertainty and studies alternative approaches [5].
References:
[1] Augustin, David, et al. "Simulating clinical trials for model-informed precision dosing: Using warfarin treatment as a use case." bioRxiv (2023): 2023-07.
[2] Augustin, David (2021). Chi - An open source python package for treatment response modelling (Version 0.1.0) [Computer software]. https://github.com/DavAug/chi
[3] Clerx, Michael, et al. "Probabilistic Inference on Noisy Time Series (PINTS)." (2019).
[4] Augustin, David, et al. "Filter inference: A scalable nonlinear mixed effects inference approach for snapshot time series data." PLOS Computational Biology 19.5 (2023): e1011135.
[5] Augustin, David, et al. "Treatment response prediction: Is model selection unreliable?." bioRxiv (2022): 2022-03.
Mathematical Modelling and Inference for Alzheimer’s Disease
Pavanjit Chaggar
Supervisors: Alain Goriely (Oxford, MI), Saad Jbabdi (Oxford ,WIN)
Industrial Supervisors:Stefano Magon (Roche), Gregory Klein (Roche)
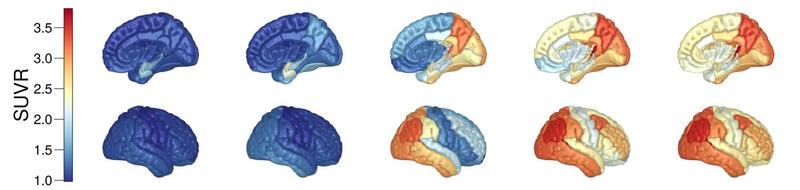
Simulated trajectory of regional tau-protein SUVR (a measure of position emission tomography tracer binding) in the human brain.
Alzheimer’s disease is a complex, multi-factorial disease about which relatively little is known. Working between Oxford and Roche, I am investigating the role of toxic proteins in driving the progression of Alzheimer’s disease. The project is very interdisciplinary, drawing mainly from methods in applied mathematic and also relying heavily on multi-modal brain imaging and statistical inference. During my PhD, I have had the opportunity to visit the Roche headquarters in Basel to learn more about their internal projects and how best to incorporate to my DPhil work into their analysis pipelines. I have also had the the opportunity to visit external collaborators and present my work internationally.
My work focusses on modelling toxic forms of tau-protein, a particularly nefarious protein that exhibits a unique spatiotemporal progression throughout Alzheimer’s disease. We have shown that simple dynamical systems embedded a brain connectivity network can describe accurately the spatiotemporal evolution of tau-protein. Furthermore, I have shown that these models can be calibrated to patient data using Bayesian inference, allowing us to investigate changes in protein dynamics across the disease timeline and make patient specific predictions that incorporate uncertainty about the observation and modelling processes.
Predicting antibody-antigen interactions using machine learning
Lewis Chinery
Supervisor: Professor Charlotte M. Deane
Industrial Supervisors: Dr Jeliazko Jeliazkov & Tejinder Bhinder (GSK)
Cancer, rheumatoid arthritis, and many infectious diseases such as SARS-CoV-2 can all be treated, to an extent, using antibody drugs. These therapeutics are incredibly powerful given antibodies’ high binding affinity and specificity to their disease targets. However, developing such treatments often takes approximately ten years and over $1 billion in investment. Speeding up and lowering the cost of this process is therefore essential to the development of more affordable, effective treatments in the coming decades.
My DPhil research focuses on developing novel computational advances to the initial ‘discovery’ stage of the drug development pipeline. This step can last approximately five years and involves testing and optimising tens of thousands of potential antibody-leads for their neutralising capability, developability, and immunogenicity. Working within the Oxford Protein Informatics Group (OPIG) in the Department of Statistics, I use advanced deep learning methods and other computational tools to enable researchers to move some of these in vitro discovery stage tests in silico, helping reduce development times and costs.
Recently, we published our antibody binding site (paratope) prediction tool – Paragraph (Chinery et al 2023. Bioinformatics). Paragraph uses the latest advances in graph neural networks to predict which amino acids of an antibody are likely to take part in binding its target. This knowledge is useful as different 3D orientations of an antibody-antigen complex can potentially lead to varying on- and off-target effects. For those wishing to better understand the methods used in this project, please take a look at our GitHub page - https://github.com/oxpig/Paragraph.
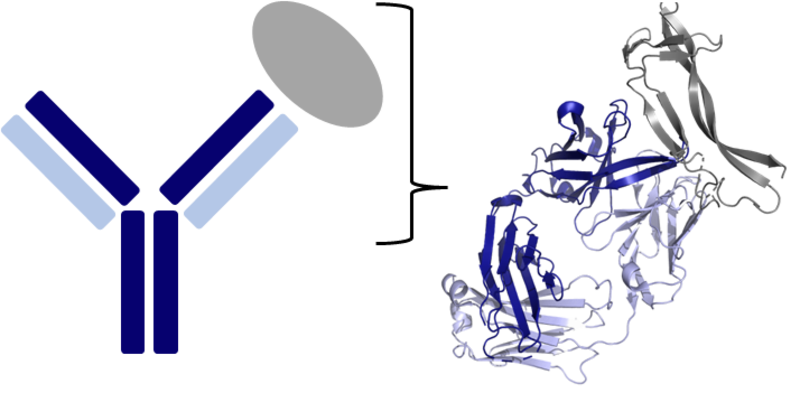
Left image shows a Y-shaped antibody bound to its target antigen (grey) e.g. a viral protein. An antibody is comprised of four protein chains – two heavy chains (dark blue) and two light chains (light blue). The right image provides a more detailed representation of one of the binding ‘arms’ of the antibody, highlighting the complex 3D conformations the protein chains form. These unique shapes allow antibodies to bind antigens with high affinity and specificity, making antibodies powerful immune proteins and therapeutics. Figure created in PyMOL.
Methods for the analysis of infant noxious-evoked potentials
Simon Marchant
Supervisors: Prof. Rebeccah Slater & Prof. Caroline Hartley
Industrial supervisor: Dr Toba Sanni (Reckitt)
Many newborn infants who are admitted to hospital undergo repeated invasive medical procedures, and there is evidence that this has both a short-term and long-term adverse impact on their neurological development. There are currently no analgesics licenced for use in newborn infants, as there is little information available about their efficacy in this population. This is partly because existing biomarkers for pain are suboptimal in neonates. The measurement of noxious-evoked potentials – the brain responses to external painful stimuli – is a promising, novel biomarker for pain which has been employed in multiple clinical trials. However, there are some limitations to the use of this method in clinical and research settings.
My work addresses these limitations. The first is contamination of the signal by external artefact: we developed a machine-learning algorithm for the automatic identification of recordings of infant noxious-evoked potential which contain artefact. The second limitation is the reproducibility of noxious-evoked potential research methods, and for this we created a standardised software pipeline for pre-processing noxious infant noxious-evoked potentials. Lastly, current methods’ lack of interpretability limit clinical use. We developed an interpretable metric for an infant’s nociceptive activity, which aims to improve the utility of this type of measure in clinical settings.
This work has real-life impact, with the potential to help infants in pain. It has produced both open-source software and a patent application.
Mathematical Modelling and Transcriptional Characterisation of Intratumoral Regulatory T Cell Sub-populations
Itai Muzhingi
Supervisors: Mark Coles and Eamonn Gaffney
Industrial supervisor: Oliver Grimm and Cristina Santini (Roche)
Regulatory T cells (Tregs) maintain immune homeostasis by suppressing harmful immune responses. However, high densities of Tregs have been shown to accumulate in solid tumours where they contribute to the suppression of anti-tumour responses. Targeting intratumoural Tregs via antibody-mediated blockade/depletion represents a promising cancer treatment strategy but remains ineffective and induces adverse events. Extensive transcriptional profiling of intratumoural Tregs and quantitative assessment of Treg migration dynamics can be applied to identify appropriate therapeutic markers and guide the development of safe and effective Treg-targeting therapeutics.
We hypothesised that tumours harbour resident and transient Tregs with distinct transcriptional and migratory profiles. To test our hypothesis, we used photoconvertible Kaede mice to label and quantify Tregs in MC38 and CT26 tumours. We developed a mathematical model to investigate whether the presence of resident and transient tumour Tregs aligns with the observed data and applied Bayesian parameter estimation to infer the rates of intratumoural Treg migration. In both tumours, we confirmed the presence of resident and transient Tregs and found that Tregs egressed from CT26 tumours to lymphoid tissue to a larger extent than MC38 tumours. Single cell analysis of intratumoural Tregs revealed that resident Tregs upregulate Lag3, Tnfrsf18 and Tnfrsf9. To investigate similarities between murine and human intratumoural Tregs, we constructed Treg meta-atlases using public single cell datasets. In mice, we observed that Lag3 and Ccr8 are unique to tumour Tregs and exhausted CD8 T cells. In humans, CCR8 expression was confined to tumour Tregs whereas LAG3 was mainly expressed on exhausted CD8 T cells.
Collectively, our findings outlined the presence of kinetically and transcriptionally distinct intratumoural Treg subpopulations. Furthermore, our Treg meta-atlases exposed key differences in CCR8 and LAG3 expression between mouse models and humans and supported the potential application of CCR8 as a target for depleting human intratumoural Tregs.
Generation of echocardiogram images for 3D image enhancement and localisation
Emmanuel Oladokun
Supervisors: Vicente Grau
Industrial supervisor: Jurica Sprem (GE Healthcare)

I experiment with generative methods based on different principles, such as adversarial, contrastive, and diffusive to create synthetic data. I then use this synthetic data to enhance existing real datasets, aiming to improve performance on tasks like left-ventricle segmentation which is key to calculating physiological variables that have diagnostic use. Moreover, the ability to localise the ultrasound probe in 3D space is becoming increasingly valuable. Therefore, I also investigate how synthetic data can be employed to train a network capable of determining the probe’s position and the viewed object, thereby significantly reducing the expertise required to perform an examination.



